Details of the Drug
General Information of Drug (ID: DM2KEW6)
| Drug Name |
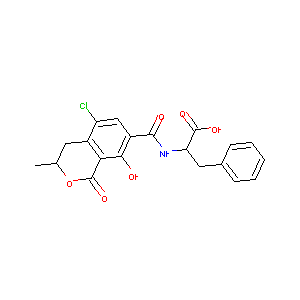
3R14S-OCHRATOXIN A
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonyms |
Ochratoxin A; OCHRATOXIN A; Antibiotic 9663; 303-47-9; Phenylalanine - ochratoxin A; NCI-C56586; C20H18ClNO6; CHEBI:7719; RWQKHEORZBHNRI-BMIGLBTASA-N; Ochratoxin A, aspergillus ochraceus; Ochratoxin A-BSA conjugate from Aspergillus ochraceus; N-(((3R)-5-Chloro-8-hydroxy-3-methyl-1-oxo-7-isochromanyl)carbonyl)-3-phenyl-L-alanine; (-)-N-((5-Chloro-8-hydroxy-3-methyl-1-oxo-7-isochromanyl)carbonyl)-3-phenylalanine; NSC 201422; UNII-1779SX6LUY; OTA; ochratoxin A
|
||||||||||||||||||||||
| Indication |
|
||||||||||||||||||||||
| Drug Type |
Small molecular drug
|
||||||||||||||||||||||
| Structure |
 |
||||||||||||||||||||||
| 3D MOL | 2D MOL | ||||||||||||||||||||||
| #Ro5 Violations (Lipinski): 0 | Molecular Weight (mw) | 403.8 | |||||||||||||||||||||
| Logarithm of the Partition Coefficient (xlogp) | 4.7 | ||||||||||||||||||||||
| Rotatable Bond Count (rotbonds) | 5 | ||||||||||||||||||||||
| Hydrogen Bond Donor Count (hbonddonor) | 3 | ||||||||||||||||||||||
| Hydrogen Bond Acceptor Count (hbondacc) | 6 | ||||||||||||||||||||||
| Chemical Identifiers |
|
||||||||||||||||||||||
| Cross-matching ID | |||||||||||||||||||||||
Molecular Interaction Atlas of This Drug
 Drug Therapeutic Target (DTT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 Drug Transporter (DTP) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Drug Off-Target (DOT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Interaction Atlas (MIA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
References
